Thanks to a multispecific sampling effort, we have been able to show that the benthic diversity of the Loyalty Islands is very rich. For a large numbers of species, the samples collected during the BIBELOT campaign complemented those of the COBELO mission (2013-ALIS), and those of a previous mission carried out on the boat of the Living Ocean Foundation (in 2013). These samples were then supplemented by those from the CHEST campaign (2015-ALIS). This sampling made it possible to estimate in these species (mainly giant clams, hydrozoans, and corals of the genus Pocillopora) the spatial scales of larval dispersal and populations connectivity.
From all these samples, several remarkable results were found in all the groups we focused on :
1) Giant clams
For the giant clams, we have demonstrated (thanks to the genetic analyzes carried out on the biopsies collected during the mission) the presence of a new species of giant clam Tridacna noae (Fig. 1), which had recently been described endemic to Taiwan (Su et al. 2014), which we found in the Loyalty Islands and the North of Grande Terre, whereas it seems to be absent from the rest of the territory (Borsa et al., 2015a, Borsa et al., 2015b). More information regarding Tridacna noae can be found in the SPC Fisheries Newsletter #145 (Wabnitz & Fauvelot 2014).

Fig.1: underwater picture of a giant clam of the species Tridacna noae, taken in Tiga (Loyalty islands, New Caledonia), during BIBELOT campaign. T. noae clams can be identified by the presence of discrete teardrops on their mantle.
Giant clams samples collected during BIBELOT has been genetically analyses, and, added to samples collected accross the Indo-Pacific, allowed to present the first phylogeny of the ten known species of the genus Tridacna, based on a multigenic analysis calibrated with fossils, in order to understand the phylogenetic relationships among species of this group and investigate their evolutionary histories. The genetic analysis of giant clam samples is still ongoing for some species (Tridacna squamosa, Tridacna derasa), but we have already shown that for Tridacna maxima, Hipopus hippopus and Tridacna noae, the populations are very structured, with limited connectivity between the populations of the Loyalty islands and those of the rest of New Caledonia (van Wynsberge et al., 2017, Fauvelot et al., 2019). Results related to this part (giant clam connectivity in New-Caledonia) can be found in a report, written in french, at this link: DOI: 10.13140/RG.2.2.28827.21286
2) Hydrozoans
For the Hydrozoans collected in New Caledonia and worldwide, the mitochondrial marker 16S was used to study lineage diversity in Aglaopheniidae and detect potential cryptic species. Additional analyses within two selected species, Lytocarpia brevirostris and Macrorhynchia phoenicea were conducted using nuclear microsatellite markers. Molecular species delimitation methods and population genetics data revealed an extensive lineage diversity and several cryptic species in the Aglaopheniidae group (Postaire et al. 2016, 2017a, 2017b). This is further illustrated, for example, in Lytocarpia brevirostris, with two lineages named apha and beta, found in sympatry on the reefs of New Caledonia (Figure 2).
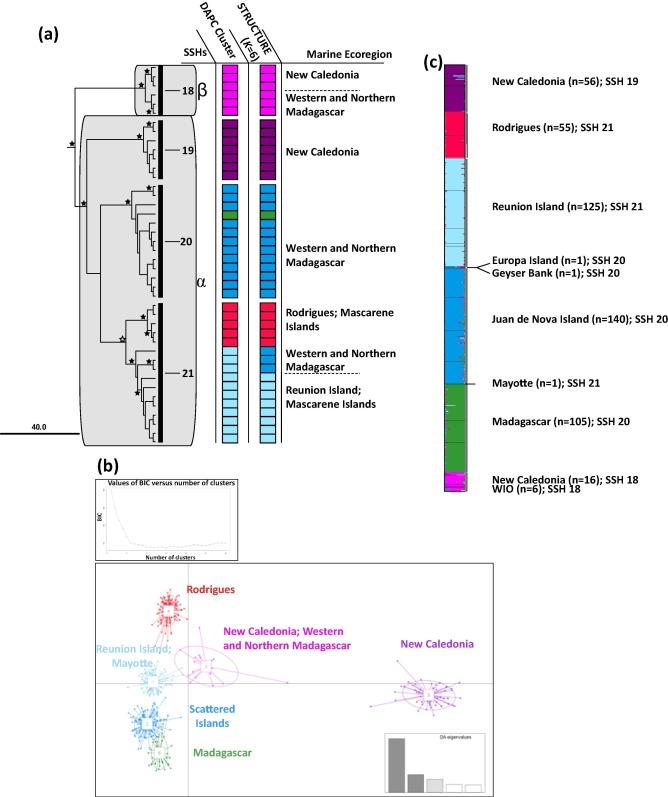
Fig. 2: (a) Sub-tree of Lytocarpia brevirostris with lineages alpha and beta and the secondary species hypotheses (SSH) resulting from the different species delimitation methods (black vertical bars) with (b) DAPC and (c) STRUCTURE outputs (Figure from Postaire et al. 2016 https://doi.org/10.1016/j.ympev.2016.08.013)
When the samples were further sorted according to their lineage membership, additional population genetic analysis showed strong structuration among sampled populations (Fig. 3), indicative of limited larval dispersal in these species (Postaire er al. 2017a, 2017b).
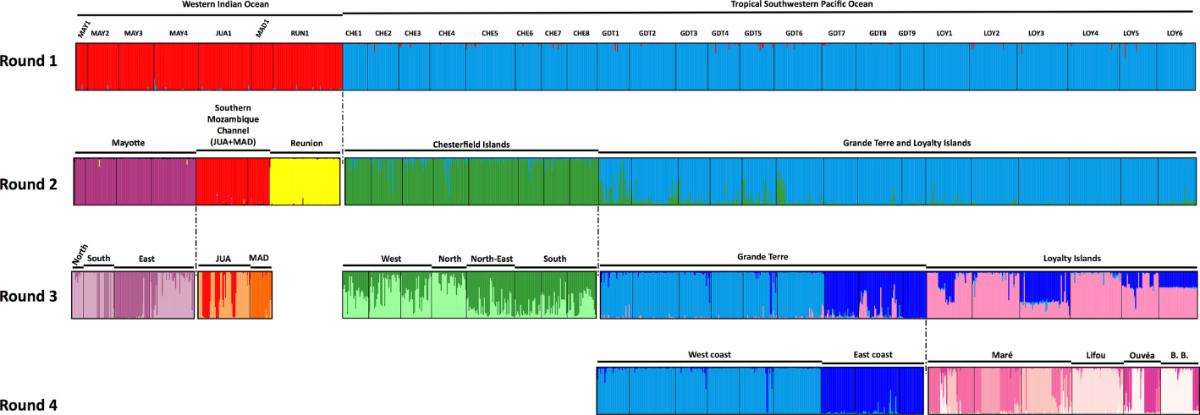
Fig. 3. Genetic population structure in Macrorhynchia phoenicea lineage alpha. Assignment probabilities of multilocus genotypes at microsatellite markers to putative clusters using an admixture model as identified by Structure. Different colors correspond to genetically differentiated clusters. Figure from Postaire et al. (2017) https://doi.org/10.1002/ece3.3236
3) Corals of the genus Pocillopora
In Pocilloporidae, results similar to those obtained in hydrozoans have been obtained, namely a number of genetically differentiated cryptic lineages, some occurring in the Loyalty Islands (Gélin et al. 2017, 2018a, 2018b). For some of the retrieved Secondary Species Hypothesis SSH within the genus Pocillopora, further genetically distinct lineages were found. This is the case of the Pocillopora eydouxi/meandrina complex sensu, Schmidt-Roach, Miller, Lundgren, & Andreakis (2014), identified by Gélin, Postaire, Fauvelot and Magalon (2017) and which was found to split into three secondary species hypotheses (SSH09a, SSH09b, and SSH09c). The SSH09b and SSH09c were almost exclusively found in the Tropical South Pacific and South Eastern Pacific (Figure 4). More surprisingly, each SSH split into two to three genetically differentiated clusters, found in sympatry at the reef scale (notably in New Caledonia, including the Loyalty Islands), leading to a pattern of nested hierarchical levels (PSH > SSH > cluster), each level hiding highly differentiated genetic groups. Thus, rather than structured populations within a single species, these three SSHs, and even the eight clusters, likely represent distinct genetic lineages engaged in a speciation process or real species. The issue is now to understand which hierarchical level (SSH, cluster, or even below) corresponds to the species one. The use (in progress) of more informative molecular markers should help us to clarify the taxonomic status of these cryptic lineages.
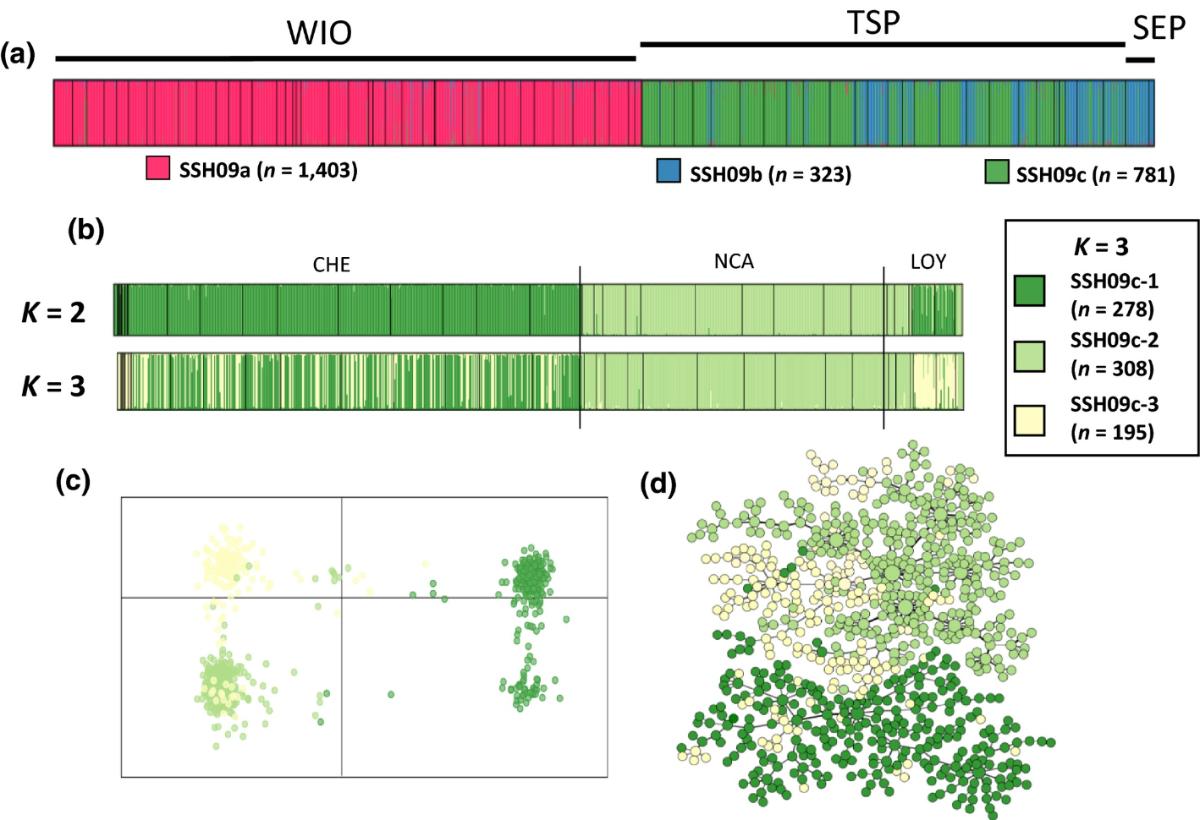
Fig. 4: Genetic variability found in the SSH09c cluster of the Pocillopora eydouxi/meandrina species complex (green cluster on Fig. 5a), using microsatellite markers. Each color corresponds to genetically differentiated clusters that correspond to divergent lineages which are found in sympatry on the reefs of Chesterfield (CHE), New Caledonia (NCA) and Loyalty Islands (LOY). (c) Results of the DAPC assignment for K = 3 and (d) the minimum spanning tree, both colored according to the three clusters identified by Structure. Figure from article available at https://doi.org/10.1002/ece3.3747
All this work represents a major advance in the field since we have been able to highlight an unsuspected number of cryptic species in this genus, which offers new perspectives for other coral genera where taxonomy based on morphological characters is still uncertain and in constant revision. Additional analyses are currently in progress in the framework of Nicolas Oury's thesis.
4) Corals other than Pocilloporidae
The other coral species were collected mainly to complete the collection of New Caledonian corals at the Noumea IRD center, but there are some new species identifications recently described in fungidae, including two symbiotic species described for the first time : a Spirobranchus richardsmithi (Pillai, 2009) serpulidae on a fungidae Podabacia motuporensis (Veron, 1990) which was the subject of a published article (Hoeksema & ten Hove 2014) and an Ancylomenes kobayashii shrimp (Okuno & Nomura, 2002) on a fungidae Polyphyllia novaehiberniae (Lesson, 1831). Also, several specimen of coral collected in the Loyalty islands have been used to complete phylogenies on hitherto unknown groups (Arrigoni et al 2016a, 2016b, 2019, Terraneo et al 2014, van der Meij et al., 2015, Waheed et al., 2015).







